
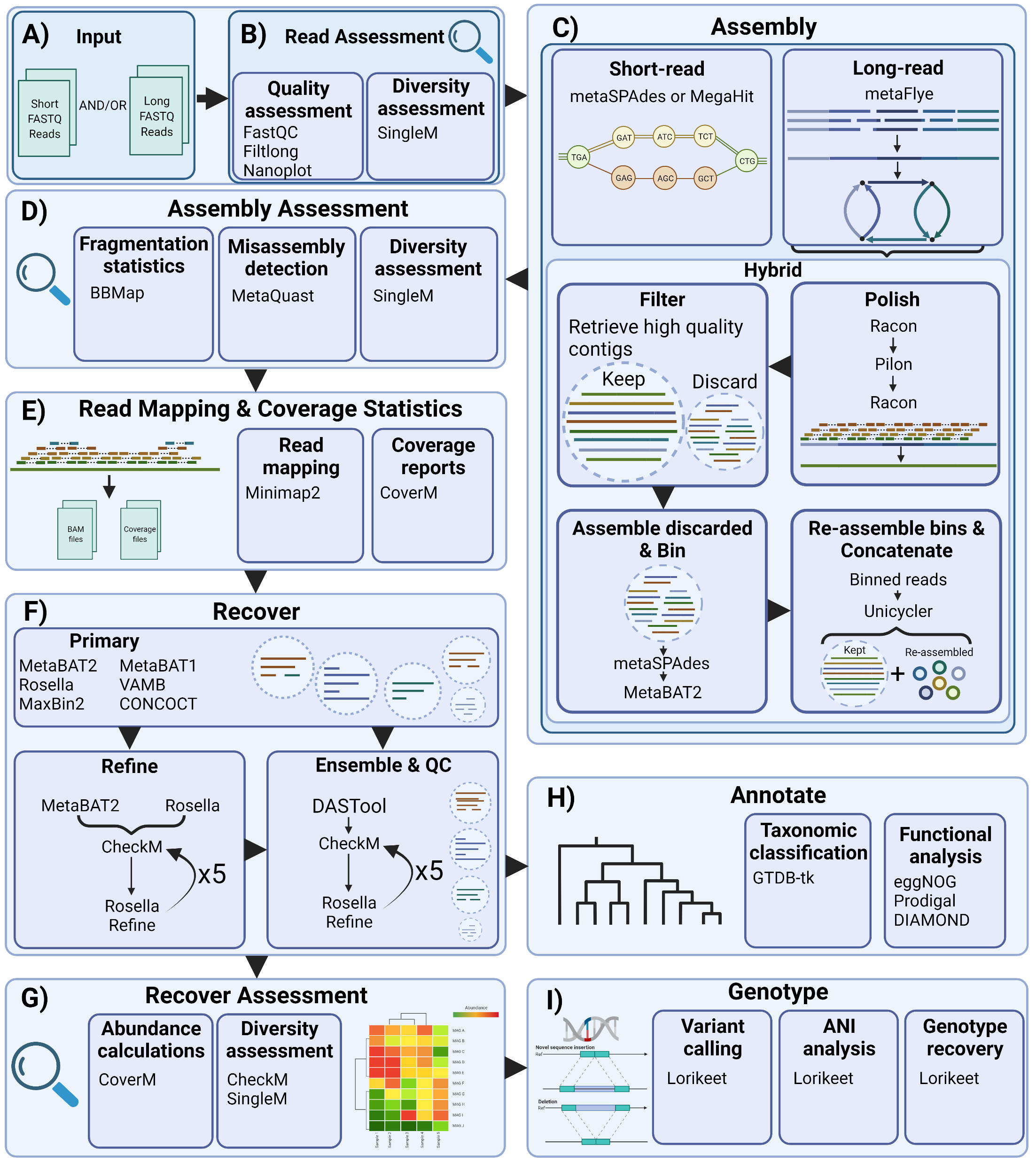
Aviary
An easy to use for wrapper for a robust snakemake pipeline for metagenomic hybrid assembly, binning, and annotation. The pipeline currently includes a step-down iterative hybrid assembler, an isolate hybrid assembler, a quality control module and a comprehensive binning pipeline. Each module can be run independently or as a single pipeline depending on provided input.
Module details
| method | description |
|---|---|
batch | Performs all steps in the Aviary pipeline on a batch file. Clusters the final output. |
cluster | Dereplicate/choose representative genomes from multiple aviary runs |
assemble | Perform quality control and assembly of provided reads. Will provide hybrid assembly if given long and short reads |
recover | Recover MAGs from provided assembly using a variety of binning algorithms. Also perform quality checks on recovered MAGs and taxonomic classification. |
annotate | Provide taxonomic and functional annotations for a given set of MAGs |
diversity | Use lorikeet to calculate strain diversity statistics on a set of MAGs and reads |
complete | Performs the complete workflow up to last possible rule given the provided inputs |
isolate | Performs hybrid isolate assembly. For use with isolated pure sequencing results. |
configure | Set or reset environment variables used by aviary |
Overview
Citation
On its way :P
License
Code is GPL-3.0
GitHub
Powered by Doctave
